The basic analysis of the data was done with the functions from Scanpy49 package (v1.9.3) of Python3 (v3.10.10) and default parameters according to Scanpy pipeline unless otherwise specified.
Mice
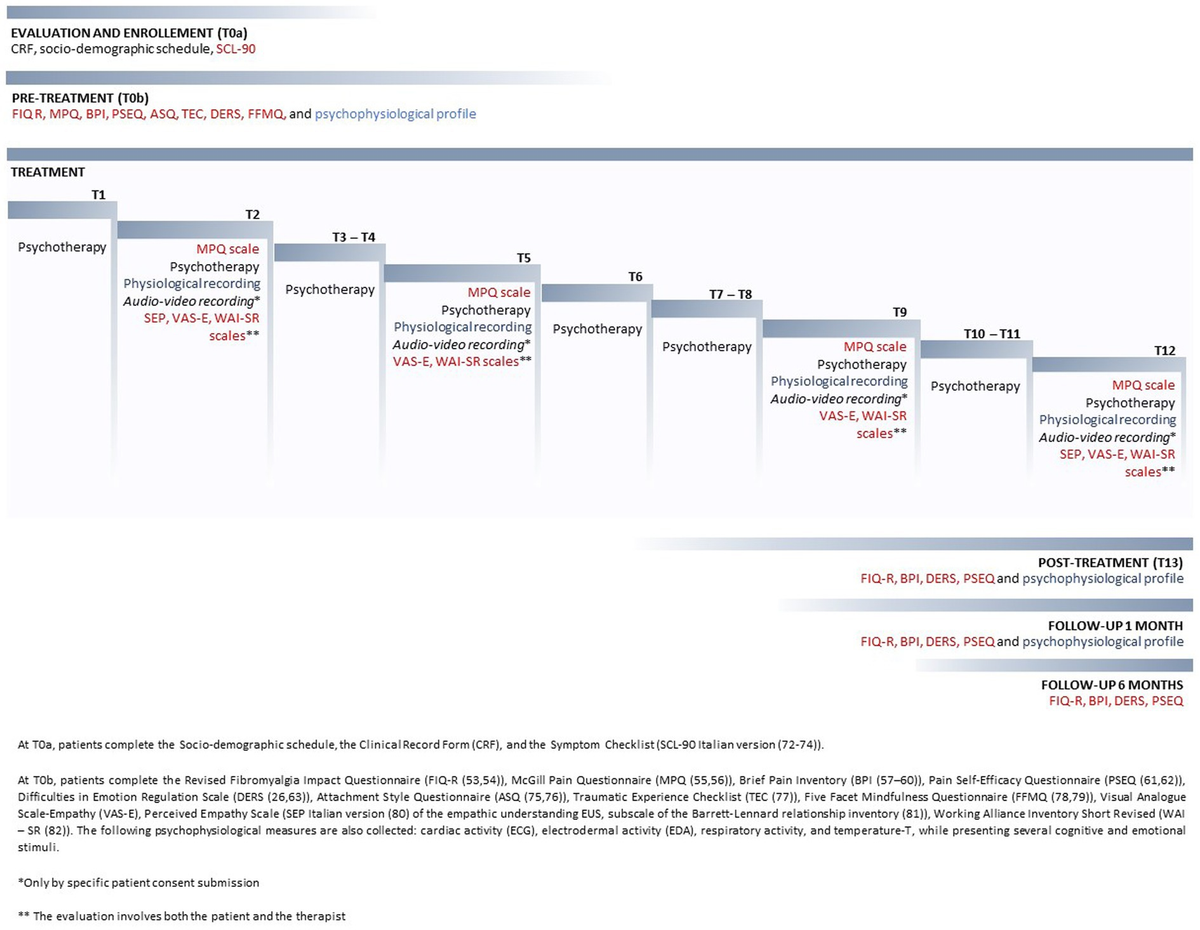
Cohorts of adult mice of both biological sexes, aged from 8 to 12 weeks were used for sequencing in all the different published studies5,9,10,11,12,13. Our sequencing data on TG and DRG were obtained from mice aged between 8 and 12 weeks using the Smart-seq3Xpress, 10x multiomic technologies, in iPain these mice are referred to as young. A middle-aged cohort (34–62 weeks) was processed using 10x multiomic sequencing, in iPain these mice are referred to as aged. For all details on biological sex, and the number of cells per timepoints see tables in Fig. S2 and the CellxGene50 iPain (iPain).
For the senescence kinetic and for the drug treatment experiment, cohorts of mice aged between 17 and 27 weeks of both sexes (3 males and 3 females per group) were used.
All the mice were on a C57BL/6 background and were kept under a 12-h light–dark cycle, at 24 °C with unlimited food and water. All animal care and experimental procedures were permitted by the Ethical Committee on Animal Experiments (Stockholm North committee) and conducted according to The Swedish Animal Agency’s Provisions and Guidelines for Animals Experimentation recommendations. Ethical permit numbers 9702-2018 and 17396-2022.
Drug administration
The senolytic compounds were given orally, by gavage. The stock solution was diluted as 10% DMSO in corn oil prior to administration. The treatment starts seven days post CCI injury, the senolytic compound or vehicle control (10% DMSO in corn oil) was administered by oral gavage once daily for either 10 consecutive days (Navitoclax CAS No. CAS. 923564-51-6 or Venetoclax CAS No. 1257044-40-8) or once weekly for 2 weeks PROTAC Bcl-xL degrader (CAS No. 2920415-08-1).
Full blood Count
Mice were deeply anesthetized with isoflurane. The full blood was taken from the hearts of the mice and kept in a blood collection tube (containing EDTA) for further testing by IDEXX BioAnalytics.
Chronic constrictive injury mice model
Chronic constriction injury of the sciatic nerve (CCI) has been widely described with minor differences in rodents51,52,53. Briefly, mice were first anesthetized with isoflurane, and a small incision was made at the mid-thigh level on the right side. Ligatures (7–0 surgical silk) were tied loosely around the sciatic nerve with approximately 0.5 mm between ligatures. And then, the skin was closed by a 5–0 silk suture.
Pain-related behavioral tests
Mechanical sensitivity was assessed by the von Frey test in an up-down testing paradigm as previously described54,55. Briefly, mice were placed in glass cylinders on a 6 × 6 mm wire mesh grid floor and were allowed to acclimate for 20–60 min. The plantar surface of the hind paw was stimulated with a series of calibrated monofilaments (von Frey hairs; Stoelting, IL) ranging between 0.008 and 2 g and withdrawal or jerking of the paw is recorded as the pain threshold. Mice were tested 6 times on each paw following the testing paradigm.
Thermal sensitivity was assessed by the Hargreaves thermal paw withdrawal test as previously described56,57. Briefly, mice were placed in Plexiglas chambers on top of a glass sheet and were allowed to acclimate for 20–60 min. A thermal stimulator (IITC Inc., Woodland Hills, CA, United States) was precisely aimed at the middle of the plantar surface of the mice through the glass sheet. The laser intensity was set to 30% and a cutoff latency of 20 s was set to avoid tissue damage.
Noxious mechanical sensitivity was assessed by pinprick test as previously described with minor modification58. Briefly, mice were placed in Plexiglas chambers on top of a glass sheet and were allowed to acclimate for 20–60 min. A 25-gauge needle connected with a 1 g filament (von Frey hairs; Stoelting, IL) was applied uniformly to the plantar surface of the hind paw without penetrating the skin. A score system was used according to the extent of the response. 0 = no response; 1 = move, look around to see what happened; 2 = brief quick lift or withdrawal or removal away of hind paw; 3 = brief quick shakes of the hind paw, or jumps; 4 = high frequency of shaking, licking, flinching, or guarding. Mice were tested 4 times on each paw with a waiting time of 5min58,59.
Anxiety and balance motor function-related behavior assessments
Anxiety was assessed by the open field test (OFT) and elevated plus maze (EPM) tests60,61. For OFT, animals were placed in a roofless 45 × 45 cm square arena enclosed with 40 cm walls and allowed to explore for 15 min while being recorded by a camera secured on the ceiling. Tracking and analysis were done using the EthoVision XT software where the arena was further partitioned into two zones: center (square area of 22.5 × 22.5 cm in the middle of the arena) and periphery (rest of arena close to walls). The proportion of time spent in the two different zones with respect to the center-point of each animal was used as an indicator of anxiety.
For EPM, animals were placed in a plus (+) shaped arena elevated at 60 cm from the ground and allowed to explore for 5 min while being recorded by a camera secured on the ceiling. Tracking and analysis were done using the EthoVision XT software where the arena was partitioned according to its design: two closed arms opposite to each other that are each 6 × 35 cm enclosed by 15 cm walls, two open arms opposite to each other with 6 × 35 cm which are all connected to one another by a small 6x6cm center area. The total distance traveled, and the proportion of time spent in the different types of zones with respect to the center-point of each animal were used as indicators of anxiety.
Motor coordination was assessed by beam walk and rotarod tests62,63,64,65. For the beam walk test, each animal and some bedding material was first placed for 2 min for habituation inside the goal box, which is 12 × 12 × 12 cm elevated to 64 cm from the ground and enclosed on all sides except for the entry side. A round beam with a diameter of 11 mm (Small) and a length of 105 cm were used. The test beam was placed straight with the endpoint in front of the goal box parallel to the ground with start and finish lines defined for a total length of 80 cm in the middle of the beam. Animals were first habituated on a 20 mm beam until the ability to fully cross the beam without hesitation was demonstrated. A total of three trials were performed on the 11 mm beam, with at least 1 min rest between trials in the goal box. Each trial was recorded on camera and the time required for the mouse to cross the beam from start to finish and the usage of the injured leg as a scoring system were used as indicators of motor function. 7 = traverses beam normally with no more than two footslips; 6 = traverses beam successfully and uses affected limb in more than 50% of the steps; 5 = traverses beam successfully and uses affected limb in less than 50% of the steps; 4 = traverses beam and places affected limb on horizontal surface at least once; 3 = traverses beam by dragging affected limbs; 2 = the animals cannot traverse but were able to place limbs on horizontal surface and maintain balance; 1 = the animal cannot traverse and were unable to put affected limb on horizontal surface.
For rotarod, the equipment from Ugo Basile S.R.L. model 47,600 was used with a start speed of 4 rpm/min, a max speed of 40 rpm, and a ramp speed of 1.8 min (20 rpm/min). Mice were placed on the rotating dowel set to minimum speed and allowed to habituate for 5 min on the first trial. A total of 8 trials were performed for each animal with a rest time of at least 5 min between each trial in the home cage. The time and final rpm of the mouse at falling or the first occurrence of passive rotation (clinging to the dowel) were used as indicators of motor function. The first 2 trials were seen as habituation trials and the last 5 trials were taken for analysis.
Senescence-associated β-galactosidase staining
Mice were deeply euthanized with isoflurane and perfused with pre-cooled PBS. L4- L6 DRGs were quickly dissected, immediately frozen, and then quickly imbedded in optimal cutting temperature embedding medium (OCT). The SA-β-galactosidase activity was detected by using a SA-β-galactosidase staining kit (Cell Signaling Technology, #9860) at PH 6.0 according to the manufacturer’s instructions with minor modification. Briefly, DRG sections were rinsed twice by PBS before fixation, followed by x-gal (PH 6.0) staining. 4 days after incubation (at 37 °C), the staining solution was removed, rinsed three times with PBS, sealed, and then immediately taken imaging.
Immunostaining
The snap-frozen DRG tissues were collected as described above (in “Methods” section, SA-β-galactosidase staining section). The snap-frozen DRG tissue was either freshly sectioned and then fixed in PFA overnight at 4 °C, or reused from SA-β-galactosidase staining (double staining). DRG sections were washed with PBS, and permeabilized with 0.3% PBST for 10 min, followed by incubation in blocking buffer (PBS containing 0.25% Triton X-100 and 1% bovine serum albumin) for 30 min at room temperature (RT). And then, DRG sections were incubated with primary antibody (diluted in blocking buffer) at 4 °C overnight. After 3 washes with blocking buffer, a secondary antibody (1:1000, diluted in blocking buffer) was applied for 1 h at RT. Cells were then incubated in DAPI solution (D212, Wako) for another 10 min, followed by 3 washes with PBS. Images were obtained with a fluorescence microscope or confocal microscope.
The following primary antibodies were used: anti-p21 antibody (1:100, gift from Doctor Sylvain Peuget), anti-peripherin (1:200; ab39374, Abcam), anti-NeuN (1:200; MAB377, Merck Millipore).
Imaging
Brightfield images were acquired using the Zeiss Axio Imager.Z2. Images were analyzed with ImageJ (v1.52 h) by (1) manual segmentation of ganglion while excluding large fibers; (2) general isolation of the blue stain with the Color Deconvolution plugin66,67 with the built-in Brilliant Blue vector; (3) thresholding by pixel intensity from 5 to 185 with Li’s method and (4) positive stain identification by the Analyze Particles function (size inclusion from 400-Infinity). Identified positive regions were refined manually before measurement of the area which were summed and normalized against the total area of the segmented DRG to determine the percentage positive area for each DRG. Statistical significance was calculated with the Mann–Whitney U test.
DRG dissociation and single-nuclei isolation51,52,53
Mice were deeply euthanized by isoflurane and perfused with 4% pre-cooled PBS. L4-L6 DRGs were collected at 14 days after CCI injury in aged mice or from young mice (without injury), and then immediately put on dry ice. Frozen tissues are stored at −80 °C before nuclei isolation.
Isolation of nuclei from frozen tissue was performed as described by the Allen Institute for Brain Science68,69. All steps were performed in 4 °C. Briefly, tissues were homogenized in 2 ml chilled homogenization buffer (10 mM Tris pH 8, 250 mM Sucrose, 25 mM KCl, 5 mM MgCl2, 0.1 mM DTT, 1× Protease inhibitor cocktail, 0.2 U/μl RNasin Plus, 0.1% Triton X-100), and filtered through 70 µm and 30 µm cell strainers. Nuclei pellets were collected by 10 min, 900 × g suspension, and then resuspended in 250 μl chilled homogenization buffer. Finally, debris was removed by Iodixanol 25–29%, and the nuclei pellet was resuspended. For single-nuclei Multiomic sequencing, the nuclei pellet was resuspended in 30 µL 1× chilled nuclei buffer (10x Genomics, 2000207) and then proceeded immediately for single-nuclei Multiomic sequencing protocol. For neuron-specific- single-nuclei RNA sequencing, the nuclei pellet was resuspended in 500 ul blocking buffer (1× PBS, 1% BSA, 0.2 U/μl RNase inhibitor) for fluorescent-activated nuclei sorting and Smart-seq3xpress.
Fluorescent-activated nuclei sorting and single-nuclei RNA sequencing (Smart-seq3xpress and 10x Multiomic sequencing)
To enable sorting of nuclei derived from neurons, nuclei suspension was incubated with anti-NeuN PE-conjugated antibody (1:500, FCMAB317PE, Merck) for 30 min on ice. Next, nuclei pellet was collected by 5 min, 400 × g suspension, and resuspended in PBS containing 0.1 μg/mL DAPI (D3571, Invitrogen) and 0.2 U/ul RNase inhibitor. Single NeuN+ and DAPI+ nuclei were sorted into each well of a 384-well plate containing 0.3 µL lysis reaction mix by a flow cytometer (DB FACSAria Fusion or BD FACSAria III) at 4 °C. Gating was performed based on DAPI and phycoerythrin signal of NeuN. After sorting, each plate was immediately spun down and stored at −80 °C before single-nuclei RNA sequencing with Smart-seq3xpress. The smart-seq3xpress protocol was performed by Rickard Sandberg’s Group, Karolinska Institute. Nuclei isolated for single nuclei Multiomic sequencing were stained with DAPI and sent for sequencing by Eukaryotic Single Cell Genomics Facility at SciLifeLab, Stockholm.
Obtaining and preprocessing the publicly available scRNA-seq data
The count matrices and associated metadata of DRG were obtained from the Gene Expression Omnibus (GEO) with the following accession numbers presented on Table 1. Additionally, the data were preprocessed according to the original papers before being loaded into Python3 as AnnData objects.
Preprocessing the lab-generated data
DRG 10x multiomics
The samples were sequenced on NovaSeq6000 (NovaSeq Control Software 1.7.5/RTA v3.4.4) with a 50nt(Read1)−8nt(Index1)−24nt(Index2)−49nt(Read2) (ATAC part) and 28nt(Read1)−10nt(Index1)−10nt(Index2)−90nt(Read2) (GEX part) setup using ‘NovaSeqStandard’ workflow in ‘S2’ mode flowcell. The Bcl to FastQ conversion was performed using bcl2fastq_v2.20.0.422 from the CASAVA software suite. The quality scale used is Sanger/phred33/Illumina 1.8+. The sequenced reads of each sample were aligned using Cell Ranger Arc pipeline version 2.0.2 to the mouse genome (mm10). After the alignment, the chromatin peaks from each sample were aggregated together to obtain a common peak set with “aggr” function from Cell Ranger Arc. The count matrices were loaded into Python3 as AnnData objects. The quality control was done by removing features that were detected in less than 3 cells, and cells with a the natural log pseudo count (log1p) of the number of genes or peaks less than 4 and more than 9 were removed. Cells with percent mitochondrial genes of more than 1.5 and a total UMI count of more than 20,000 were also filtered out.
Smart-seq3 DRG & TG
The sequenced reads were aligned using the zUMIs pipeline which makes use of the STAR-SOLO70 alignment tool, and the reference genome was mouse (mm39). After alignment, the count matrices were loaded into Python3 for analysis. Furthermore, the count matrices were filtered to only yield high-quality cells. For DRG, genes found in less than 20 cells, and cells with less than 1000 genes were filtered out. For TG, genes detected in less than 3 cells, and cells with more than 3 percent of mitochondrial genes and less than 2000 detected genes were removed.
Main UMAP of DRG
The AnnData objects were concatenated to create a unified AnnData object, and we made use of two models which were based on the scVI71 framework (v0.20.3) for the integration of DRG data. First, we trained the scVI model to integrate the DRG cells unbiasedly on 3000 highly variable genes. The model architecture was designed to obtain 50 dimensions in the latent space with 5 hidden layers and 128 nodes per hidden layer, where the distribution of gene likelihood was negative binomial. Then we constructed a scANVI model based on the trained scVI model to obtain the latent space that could clearly separate the respective cell types for visualization. We used the cell-type nomenclature from Renthal et al. 5 to train the model and perform label transfer. The scANVI model was trained until the model was converged. After the models were trained, the latent spaces from scVI and scANVI were extracted. The neighbor graph was built from 50 dimensions with 100 neighbors and the method to calculate the distance was cosine based on the latent space from scANVI. The projection of UMAP of constructed from the neighbor graph with parameter min_dist = 0.5 before plotting.
Feature imputation
Next, we train the MultiVI model to imputed for the missing features across data modalities by using our lab-generated multiomics data as an anchor. We aimed to impute as many features as possible. The MultiVI was constructed by using the default parameters and was trained until it converged. To train the model, we employed NVIDIA A100 GPU to be able to hold the count matrix on the VRAM, and this resource was provided by the National Academic Infrastructure for Supercomputing in Sweden (NAISS). After the training, the imputation of the gene expression modality was done by calling get_normalized_expression method from the model, while the chromatin accessibility modality was imputed using get_accessibility_estimates method with custom parameters of threshold = 0.1, normalize_cells = True, and normalize_regions = True. The imputed matrices were then multiplied by 10,000.
Analysis of the nociceptive lineage cells
The nociceptive lineage cells were selected and subset to yield high-resolution clusters. The neighbor graph and UMAP projection were calculated using the same parameters as mentioned above. Then we clustered the nociceptive lineage cells with the Leiden clustering algorithm at 0.1 resolution. To visualize the distribution of cells along the time kinetic after injury, the embedding_density method from the Scanpy package was utilized to compute the cell density on the UMAP projection for cells at different timepoints. Next, we ran the pySCENIC72 pipeline to calculate the regulon activity in the nociceptive lineage. Yet, not all TFs had the information for regulons and thus we fit the univariate linear model (ULM) from the decoupleR73 package by using the RNA expression of each cell as an independent variable and the interaction weight of each TF from the adjacency table from pySCENIC as a target. The t-value of the slope of the fitted model represented the TF activity.
Cell state potency
The relative cell state potency of the nociceptive lineage cells was calculated in R using the CytoTrace package. Before running CytoTrace, we reconstructed the raw count matrix of the gene expression data from the imputed normalized count with the PyTorch distribution module for the negative binomial distribution. After the reconstruction, the AnnData object was converted into a SingleCellExperiment (SCE) object using rpy2 and anndata2ri Python packages. Once converted, we ran CytoTrace on the reconstructed raw count matrix with data sources passed to the batch parameter. The CytoTrace was normalized to have the range from 0 to 1. Then we calculated the CytoTrace pseudotime by taking the difference between 1 and the normalized CytoTrace score. We performed an independent t-test with 7000 random permutations on the CytoTrace scores between the naïve and injury states with the SciPy package to confirm that the score in the naïve state was significantly higher than that of the injured state.
Temporal analysis of nociceptive subtypes
We employed CellRank74 (v2.0.0) to perform the temporal analysis of each nociceptive subtype between control and pain states based on RNA modality. For each subtype, we made use of the CytroTrace kernel from CellRank to compute the transition matrix. Then we passed the transition matrix CytroTrace kernel to Generalized Perron Cluster-Cluster Analysis (GPCCA) for the identification of terminal states and fate probability toward the terminal states with n_states=3. With GPCCA, we identified the lineage driver genes that drove the transition of cells toward the terminal state of pain. A generalized additive model was utilized to fit the expression profile for each gene as a function of pseudotime to visualize the expression trend of the lineage driver genes. Furthermore, we also applied the same analytical approach on the regulons level calculated from SCENIC.
Cell-cell communication
The inference of cell-cell communication (CCC) was done with the LIANA+75 package (v0.1.8). Here, we passed the count matrix of the atlas as input to the rank_aggregate.by_samplen function of LIANA and we selected “mouseconsensus” as the resource for CCC inference. Due to multi-conditions and multi-timepoints information held within the atlas, the communications were very complex. Thus, tensor decomposition was employed to extract meaningful communications as factors. This was accomplished by calling run_tensor_cell2cell_pipeline function provided by the cell2cell28 package.
Identification of senescence
The SenMayo gene score was calculated based on the scaled data of the imputed gene expression count matrix by using the score_genes function from the Scanpy package. The SenMayo gene set can be found in Supplementary Data 7. Additionally, we selected cells within the top 10 percentile and assigned them as SASP. To study the relative age of SASP cells, we performed gene set enrichment analysis with the run_gsea function from the decoupleR73 package to extract the enrichment scores of GenAge and CellAge gene sets. This was done on the normalized unscaled imputed gene expression count matrix. Mann–Whitney U test was performed to test for the statistical significance of the SenMayo score between the naïve and injury groups.
Analysis of human DRG RNA-seq within
For the analysis of human DRG RNA-seq, we first obtained the bulk RNA-seq data of different pain conditions of neuron-rich samples from Ray et al.38. As the count matrix was normalized using the TPM method, we multiplied 1e6 to the matrix to unnormalize it back to the raw transcript count values. Then we normalized the raw count to obtain the total transcript count per sample after normalization equal to the median of the total transcript count for that sample before normalization. The normalized count matrix was log-transformed and scaled with log1p and scale functions from Scanpy. SenMayo score was calculated with the score_gene function on the scaled count matrix.
Regarding the deconvolution of hDRG, we retrieved the snRNA-seq of hDRG under normal conditions from Jung et al.39 with the accession number GSE201654 from GEO. The data was preprocessed and analyzed as described in the original paper. The deconvolution was done using a VAE-based model called Bulk2Single provided by the omicverse76 package in Python3. The bulk RNA-seq data of hDRG was separated into two count matrices, one from donors without known pain conditions and the other from donors with pain. As the data was split in two, we trained two models separately for 3500 epochs each. After the training, the predicted snRNA-seq count matrices were reconstructed and merged for further analysis. There were more than 250,000 cells generated by the models in total. We processed and clustered the matrix according to Scanpy workflow with the Leiden clustering algorithm, and we noticed that there was a lot of noise within the generated data as there were many clusters comprised of 50–1000 cells. Thus, Leiden clusters with less than 900 cells were removed to retain cells with clean signals. Furthermore, we selected only nociceptive cells to calculate the SenMayo score after scaling the count matrix.
Regarding the analysis of hDRG, we noted that the cell proportions between the reference snRNA-seq and the deconvolution of bulk RNA-seq were not equal. This result was expected as we performed the deconvolution on the samples from the group being classified by the authors as neuron-rich samples36 (51 out of 70 samples), to focus our analysis on nociceptive lineage cells. The deconvolution yielded ~60% neurons whereas the reference snRNA-seq contained a more representative population of the cell types in DRG with around ~2% neurons. We thus applied a broad classification on the hDRG in contrast to the single-cell analysis in mice model, as we suspected that the deconvolution model could not perform well in classifying the high-resolution cell types (neuronal subtypes) that were closely related to each other due to it being trained on the reference sample with lower neuron representation. Nevertheless, even with the broad classification the model seems to have some difficulties in clearly distinguishing between the two neuronal cell types of NFs and nociceptors that are more highly correlated than other cell types (Fig. 4h). Despite this, the Spearman correlation still indicated the highest correlation between the transcriptome profiles of the nociceptors identified in the reference snRNA-seq and the deconvolved RNA-seq nociceptors, indicating the applicability of the model. We then proceeded to perform a SenMayo score comparison between the NoPain and Pain groups to find that it is increased in the Pain group indicating for cellular senescence, aligning with our other results.
Analysis of human DRG RNA-seq with diabetic neuropathy
The count matrix was obtained from Hall et al.40. The count matrix was normalized into transcript per million and pseudo-log transformed. The effect of different sexes was regressed out with Scanpy’s regress_out function. Samples with their ages being outliers between the healthy and diabetic groups were removed; these were samples with ages below 30 and ages above 60. The SenMayo score was calculated after the matrix was being scaled. The Z-score of SenMayo was computed before performing a one-sided t-test to statistically compare the scores between healthy and diabetic groups.
UMAP of TG
For the TG, we generate an integrated atlas separately for TG using scGLUE20 (v0.3.2), by first computing the latent space representation of each source and modality and passing them as input. For the latent space representation of RNA, we updated the weights of the scANVI model trained on the DRG data to obtain the updated latent space of the RNA gene expression modality for the TG datasets. LSA was performed on the ATAC modality to obtain the latent semantic indexing from the TG datasets. Furthermore, scGLUE required an additional input besides the latent space representations which was the guidance graph. The guidance graph was constructed using the highly variable features from RNA and ATAC as described in the original paper20. After the inputs were prepared, the model was trained with the default parameters. Once the model was trained, we extracted cell embedding from the model to build a neighbor graph using “cosine” as a metric. The UMAP was computed from the neighbor graph. To facilitate us with cell-type annotation, we perform label transfer using the scANVI model that was previously updated with the weights.
Statistical analysis
The statistical analysis and tests were done in Python3 with the “stats” module from the SciPy v1.10.1 package. The data were either imported as DataFrame or Array through Pandas v2.0.3 or Numpy v1.22.4 package respectively. In general, a t-test was applied when the data followed a normal distribution. Otherwise, the Mann–Whitney U test was performed if the data did not appear to follow the normal distribution. For the behavior analysis with paired data paired t-test was used for comparison, where an independent t-test was used on unpaired data.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.